A Method is developed for validating the Accuracy of NMR Protein Structures
– Expected to accelerate drug discovery
A team of collaborative international researchers, Adnan Sljoka at RIKEN Center of Advanced Intelligence Project (AIP), have developed a computational method named ANSURR (Accuracy of NMR structures using Random Coil Index and Rigidity) to measure the accuracy of protein structures solved by experimental technique Nuclear Magnetic Resonance (NMR).
The outcome of this research is expected to accelerate and improve the process of protein structure determination and aid in development of new therapeutics.
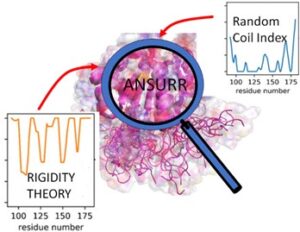
Figure: ANSURR evaluates the accuracy of NMR protein structure by comparing two measures of protein flexibility (orange predicted from structure using mathematical Rigidity Theory and blue derived from Random Coil Index using experimental NMR chemical shift data).
Proteins are fundamental biological molecules for all living organisms with essential role in myriad of biological processes. Accurate representation of their 3-dimensional structures at atomistic level is critical for researchers to understand how proteins function and in drug discovery applications.
Majority of protein structures have been solved by X-ray Crystallography or NMR experimental technologies. NMR detects the magnetic properties of molecules to identify their arrangement in space which has the advantage over X-ray crystallography as it can determine structures of proteins in solution (in its natural environment) without crystallizing the protein sample. Structure determination by NMR is crucial as many proteins cannot be crystallized. Solved protein structures are model representation of experimental data which have uncertainties that are prone to measurement and modelling errors. In contrast to crystal structures, which have well-established methods for assessing their structural accuracy, validating the quality of NMR structures has been extremely difficult. Since the first protein was published using NMR by Williamson (co-author of this study) and others in 1985, until now there has been no effective method for NMR protein structural validation, which has largely limited their applications and use among protein researchers.
In this study, the authors succeeded in developing a method ANSURR for evaluating the accuracy of NMR protein structures. ANSURR is based on comparison of two independent metrics for quantifying local protein flexibility for each amino acid in the protein. One metric is based on a measure of protein flexibility using experimentally derived NMR chemical shift data (a data collected with NMR but not used in structure determination) which is utilized in random Coil Index (RCI) and (ii) second is based on techniques from mathematical rigidity theory previously developed by Sljoka and others which predicts flexibility using protein structures as inputs. For a given NMR structure, if the correlation between RCI and rigidity theory predictions of flexibility is high, the quality of NMR structure is determined by ANSURR to be high. Rigidity theory is an area of discrete mathematics that is concerned with determining rigidity and flexibility properties of man-made structures, such as bridges, buildings and mechanical linkages in robotics, or natural structures such as zeolites and molecules, which has important applications in analysis of protein functions.
The authors had validated and extensively tested the performance of their method on both crystal and NMR structures. Development of ANSURR is a major advancement in long-standing problem in protein structure validation. It is expected that this method will give researchers more confidence in the use structural data from NMR and for scientists involved in structure determination process. The future goal is to evaluate the accuracy of all NMR structures in Protein Data Bank. This work is expected to open new research avenues in protein structure determination and improve the standard for protein structure refinement. Ultimately this should lead to better understanding how proteins perform their function, with general implications for structural biology research. This research has exemplified how mathematics and algorithms can provide significant contributions to structural biology and fundamental biological applications.
This research is published in journal Nature Communications (December 18, 2020).
<DOI> 10.1038/s41467-020-20177-1
Title of paper: A method for validating the accuracy of NMR protein structures
Authors:
RIKEN Center for Advanced Intelligence Research
Adnan Sljoka
University of Sheffield
Nicholas J. Fowler and Mike P. Williamson
For more details see press release (in Japanese)




